Fitting a real-data covariance matrix: interpretation
Jason Willwerscheid
9/11/2020
Last updated: 2020-09-17
Checks: 6 0
Knit directory: drift-workflow/analysis/
This reproducible R Markdown analysis was created with workflowr (version 1.2.0). The Report tab describes the reproducibility checks that were applied when the results were created. The Past versions tab lists the development history.
Great! Since the R Markdown file has been committed to the Git repository, you know the exact version of the code that produced these results.
Great job! The global environment was empty. Objects defined in the global environment can affect the analysis in your R Markdown file in unknown ways. For reproduciblity it’s best to always run the code in an empty environment.
The command set.seed(20190211) was run prior to running the code in the R Markdown file. Setting a seed ensures that any results that rely on randomness, e.g. subsampling or permutations, are reproducible.
Great job! Recording the operating system, R version, and package versions is critical for reproducibility.
Nice! There were no cached chunks for this analysis, so you can be confident that you successfully produced the results during this run.
Great! You are using Git for version control. Tracking code development and connecting the code version to the results is critical for reproducibility. The version displayed above was the version of the Git repository at the time these results were generated.
Note that you need to be careful to ensure that all relevant files for the analysis have been committed to Git prior to generating the results (you can use wflow_publish or wflow_git_commit). workflowr only checks the R Markdown file, but you know if there are other scripts or data files that it depends on. Below is the status of the Git repository when the results were generated:
Ignored files:
Ignored: .DS_Store
Ignored: .Rhistory
Ignored: .Rproj.user/
Ignored: data/.DS_Store
Ignored: data/datasets/
Ignored: docs/.DS_Store
Ignored: docs/assets/.DS_Store
Ignored: output/
Untracked files:
Untracked: analysis/extrapolate3.Rmd
Untracked: analysis/extrapolate4.Rmd
Untracked: analysis/pm1_priors_tmp.Rmd
Unstaged changes:
Modified: analysis/index.Rmd
Modified: analysis/tree_literature.Rmd
Modified: drift-workflow.Rproj
Note that any generated files, e.g. HTML, png, CSS, etc., are not included in this status report because it is ok for generated content to have uncommitted changes.
These are the previous versions of the R Markdown and HTML files. If you’ve configured a remote Git repository (see ?wflow_git_remote), click on the hyperlinks in the table below to view them.
| File | Version | Author | Date | Message |
|---|---|---|---|---|
| Rmd | 0c9b4ed | Jason Willwerscheid | 2020-09-17 | wflow_publish(“analysis/covmat_1kg_shared3.Rmd”) |
suppressMessages({
library(flashier)
library(drift.alpha)
library(tidyverse)
})I interpret results from the previous analysis. I do a bimodal drift fit and then sort factors by the L2 norm of their loadings.
Superpopulations
covmat <- readRDS("../data/datasets/1kg_phase3_derived/1kg_phase3_derived_covmat.rds")
meta <- readRDS("../data/datasets/1kg_phase3_derived/1kg_phase3_derived_meta.rds")
plot_fl <- function(LL) {
df <- data.frame(LL)
colnames(df) <- paste0("Factor ", formatC(1:ncol(LL), width = 2, flag = "0"))
df$subpop <- meta$pop
df$superpop <- meta$super_pop
df <- df %>% arrange(superpop, subpop)
df$idx <- 1:nrow(df)
gath_df <- df %>%
gather(K, value, -subpop, -idx, -superpop) %>%
mutate(K = factor(K))
med_gath_df <- gath_df %>%
group_by(subpop, K) %>%
summarise(value=median(value), idx=median(idx))
p <- ggplot(gath_df, aes(x=idx, y=value, color=superpop)) +
geom_point() +
facet_wrap(~K) +
geom_hline(yintercept = 0, linetype = "dashed") +
theme(axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank()) +
labs(color="superpop")
return(p)
}
plot_subpops <- function(LL) {
df <- data.frame(LL)
colnames(df) <- paste0("Factor ", formatC(1:ncol(LL), width = 2, flag = "0"))
df$subpop <- meta$pop
df$superpop <- meta$super_pop
df <- df %>% arrange(superpop, subpop)
df$idx <- 1:nrow(df)
gath_df <- df %>%
gather(K, value, -subpop, -idx, -superpop) %>%
mutate(K = factor(K))
med_gath_df <- gath_df %>%
group_by(subpop, K) %>%
summarise(value=median(value), idx=median(idx))
all_plots <- lapply(levels(df$superpop), function(pop) {
p <- ggplot(filter(gath_df, superpop == pop), aes(x=idx, y=value, color=subpop)) +
geom_point() +
facet_wrap(~K) +
geom_hline(yintercept = 0, linetype = "dashed") +
theme(axis.title.x=element_blank(),
axis.text.x=element_blank(),
axis.ticks.x=element_blank()) +
labs(color="subpop") +
ggtitle(paste("superpop:", pop))
return(p)
})
}t_greedy <- system.time({
ones <- matrix(1, nrow = nrow(covmat), ncol = 1)
ls.soln <- t(solve(crossprod(ones), crossprod(ones, covmat)))
covmat_diagNA <- covmat
diag(covmat_diagNA) <- NA
fl_g <- flash.init(covmat_diagNA) %>%
flash.set.verbose(0) %>%
flash.init.factors(EF = list(ones, ls.soln)) %>%
flash.fix.loadings(kset = 1, mode = 1) %>%
flash.backfit() %>%
flash.add.greedy(Kmax = 11,
prior.family = prior.point.laplace())
})
cat("Time to fit greedy factors:", round(t_greedy[3], 1), "seconds")#> Time to fit greedy factors: 41.9 secondst_backfit <- system.time({
fl_bf <- fl_g %>% flash.backfit(maxiter = 1000)
})
cat("Time to backfit:", round(t_backfit[3] / 60, 1), "minutes")#> Time to backfit: 35.7 minutesLL <- fl_bf$loadings.pm[[1]] %*% diag(sqrt(fl_bf$loadings.scale))
LL <- LL[, c(1, rep(2:fl_bf$n.factors, each = 2))]
LL <- t(t(LL) * c(1, rep(c(1, -1), fl_bf$n.factors - 1)))
LL <- pmax(LL, 0)
LL_scale <- apply(LL, 2, max)
LL <- t(t(LL) / LL_scale)
t_drift <- system.time({
dr <- init_from_covmat(covmat, LL, p = 10000, prior.s2 = LL_scale^2, fix.EL = 1,
prior.family = prior.bimodal())
dr <- drift(dr, maxiter = 500, tol = 1e-4, verbose = FALSE)
})
cat("Time to drift:", round(t_drift[3] / 60, 1), "minutes")#> Time to drift: 11.9 minutes
LL <- dr$EL %*% diag(sqrt(dr$prior_s2))
LL_norms <- apply(LL, 2, function(x) sum(x^2))
LL <- LL[, order(LL_norms, decreasing = TRUE)]
LL_norms <- apply(LL, 2, function(x) sum(x^2))
zero_cols <- which(LL_norms < 1e-6)
LL <- LL[, -zero_cols]
plot(plot_fl(LL))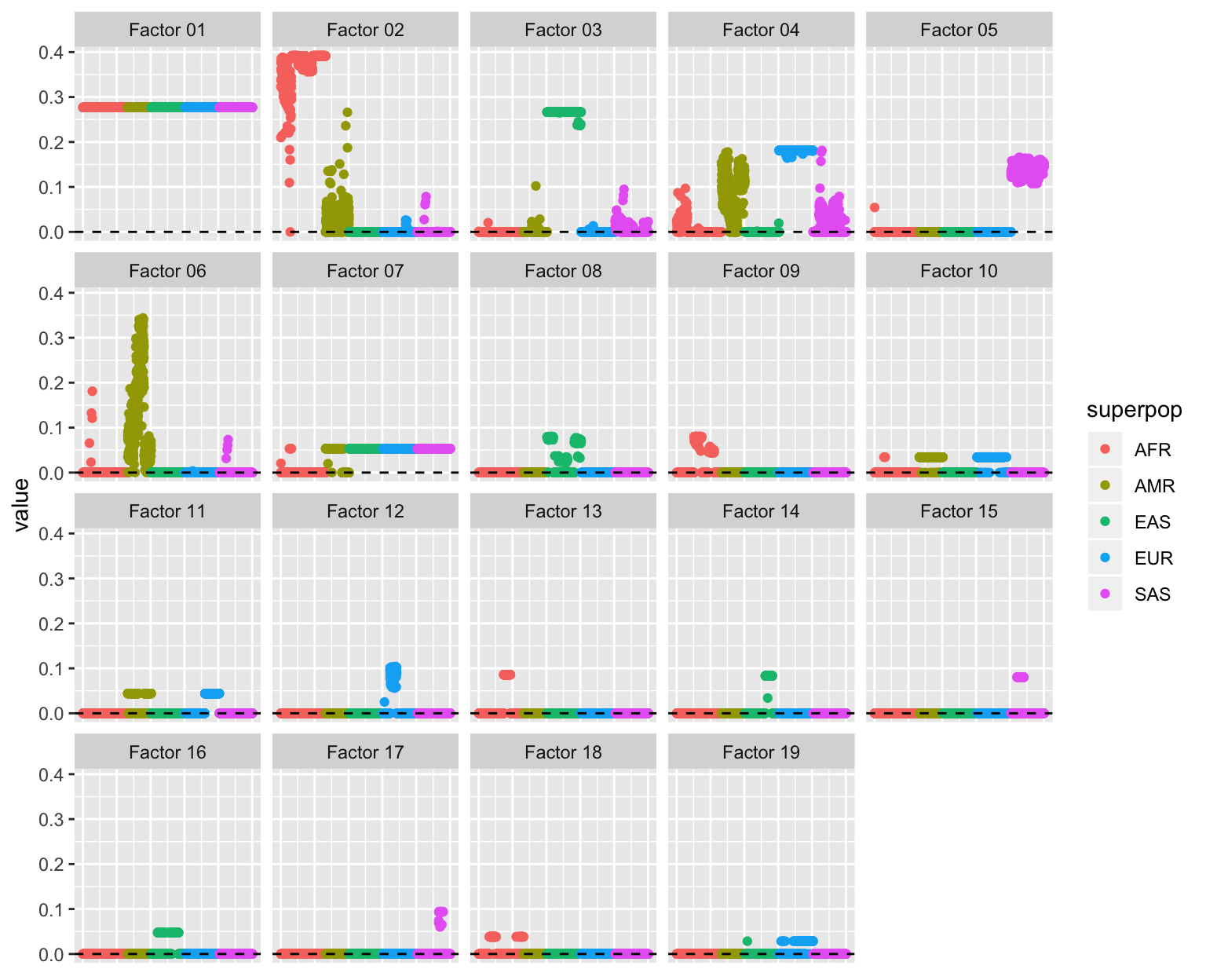
Subpopulations
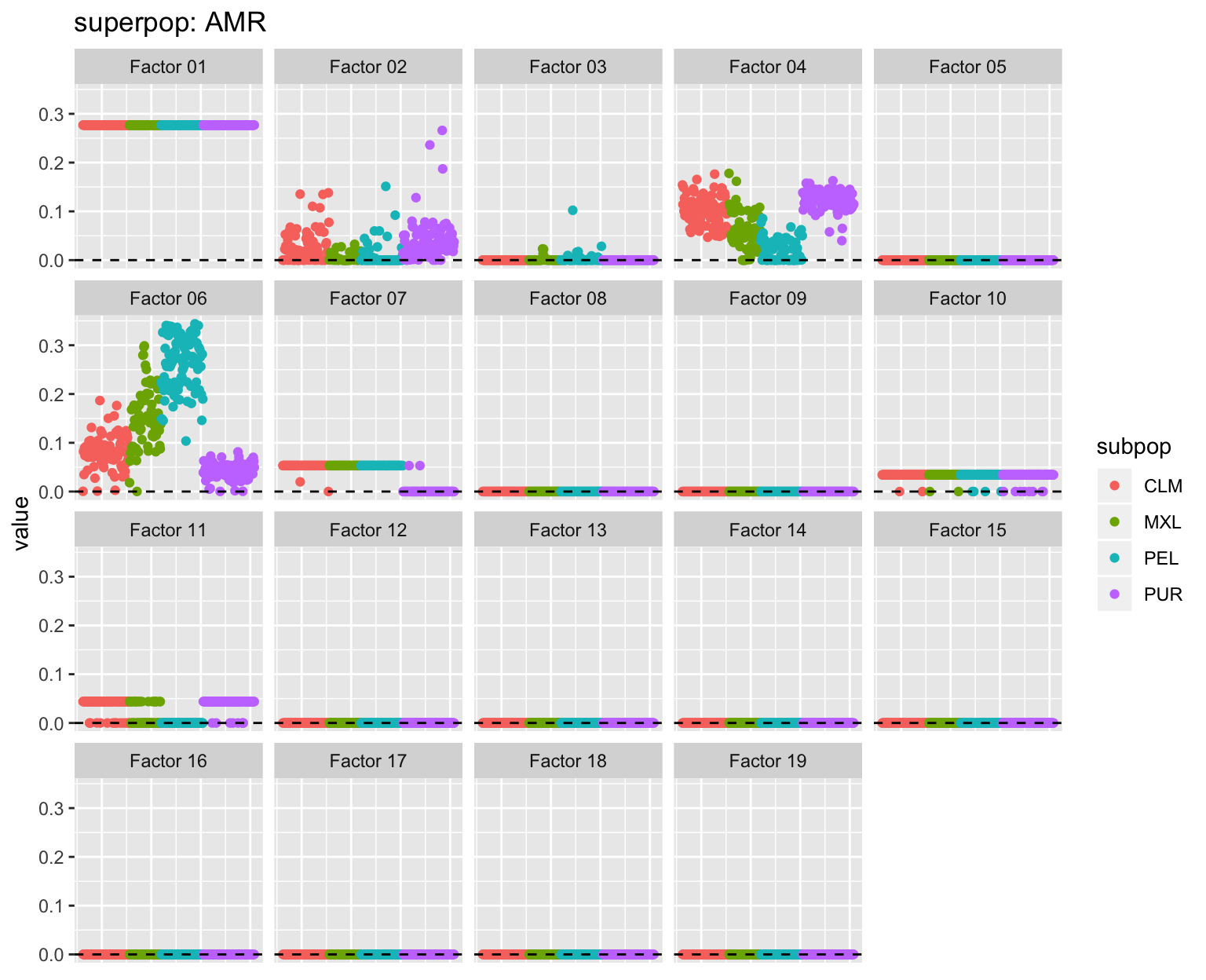
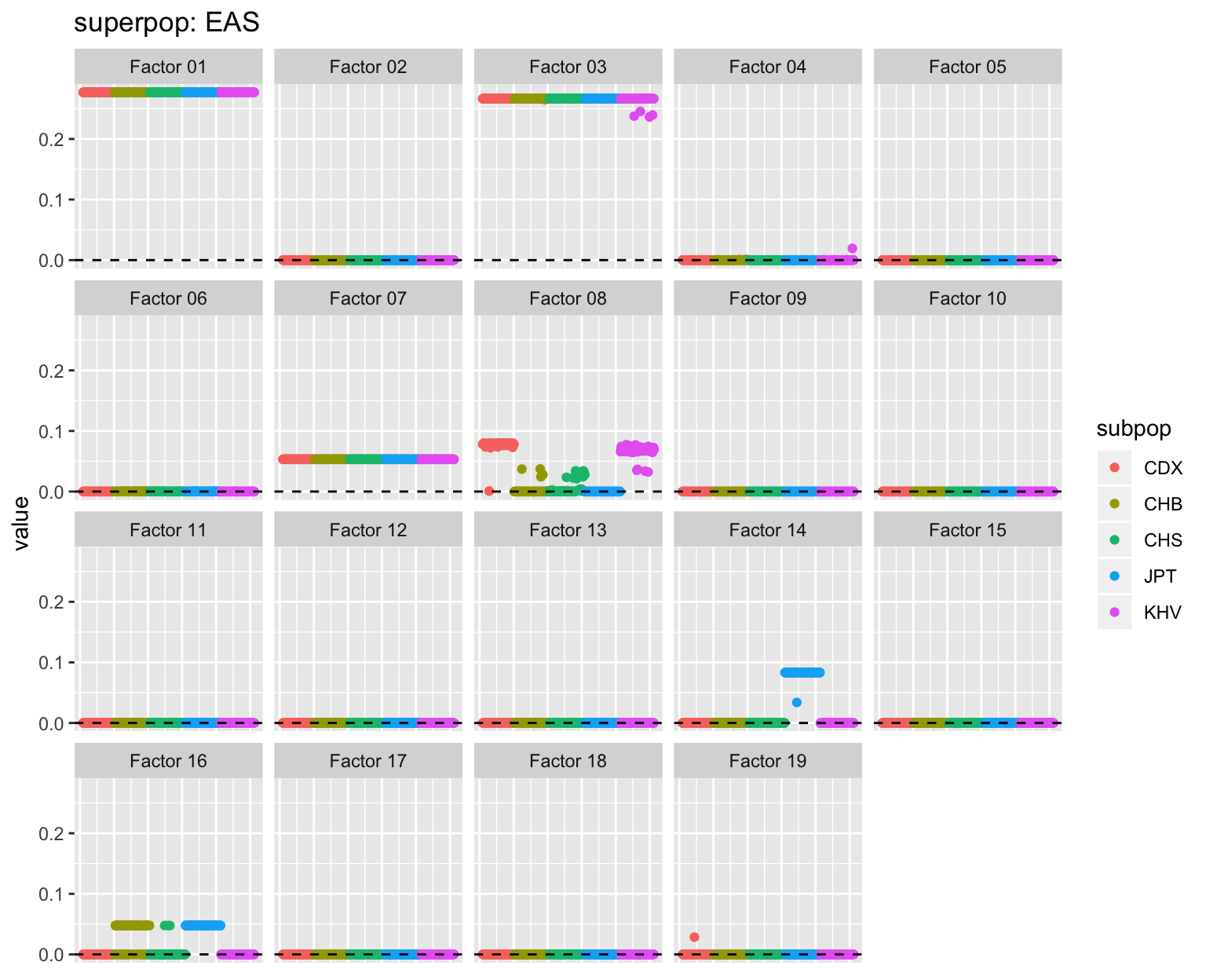
all_p <- plot_subpops(LL)
for (p in all_p) {plot(p)}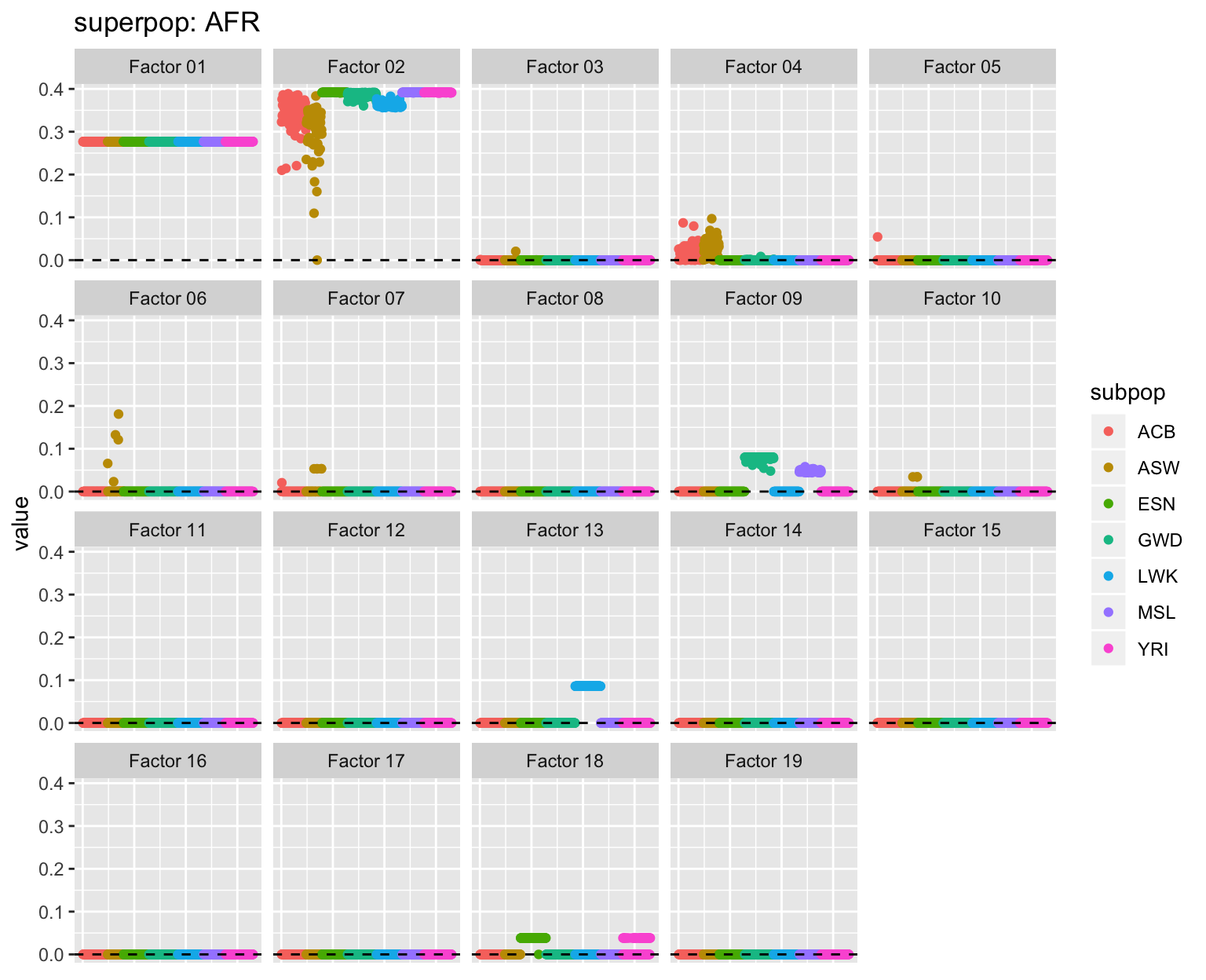
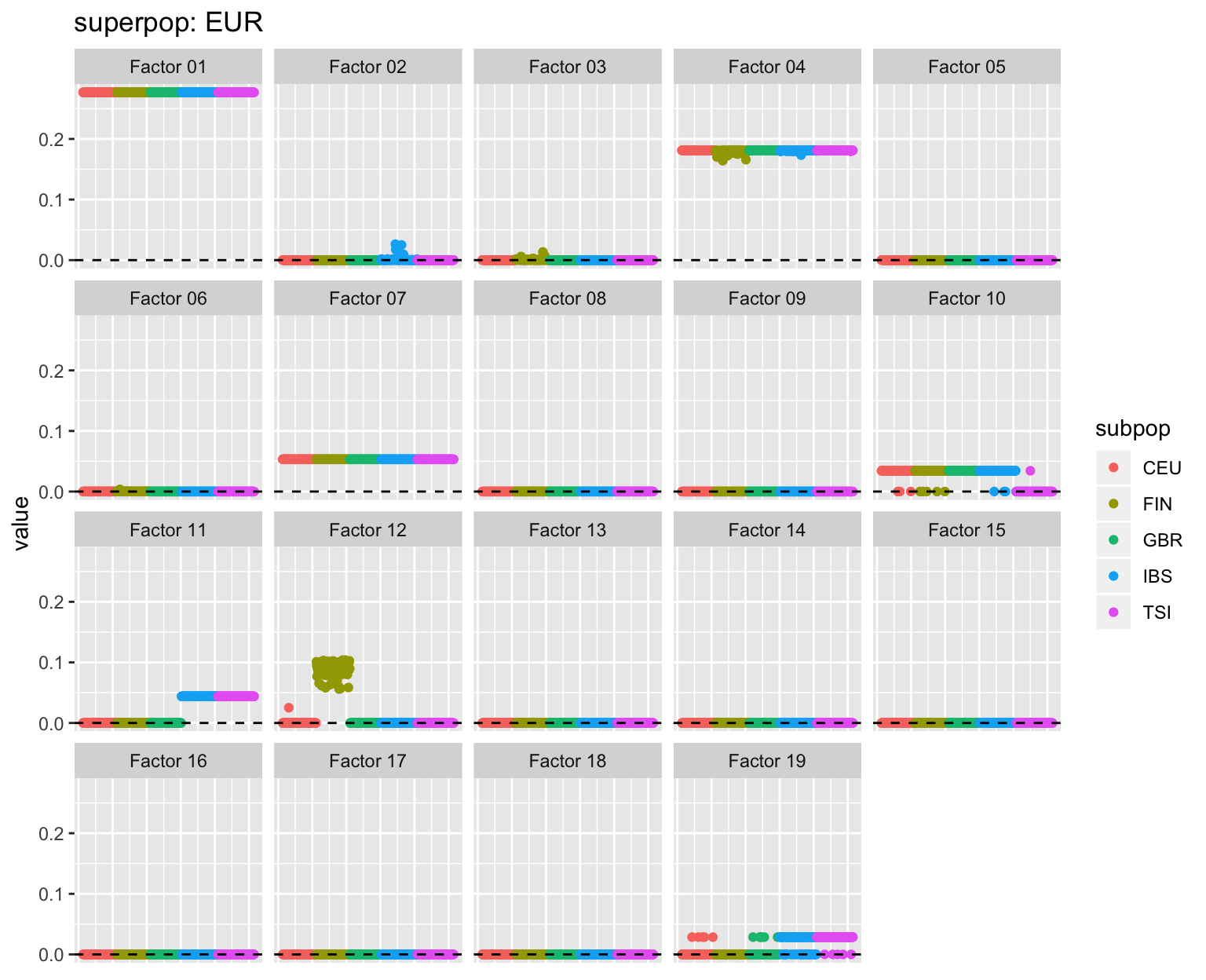
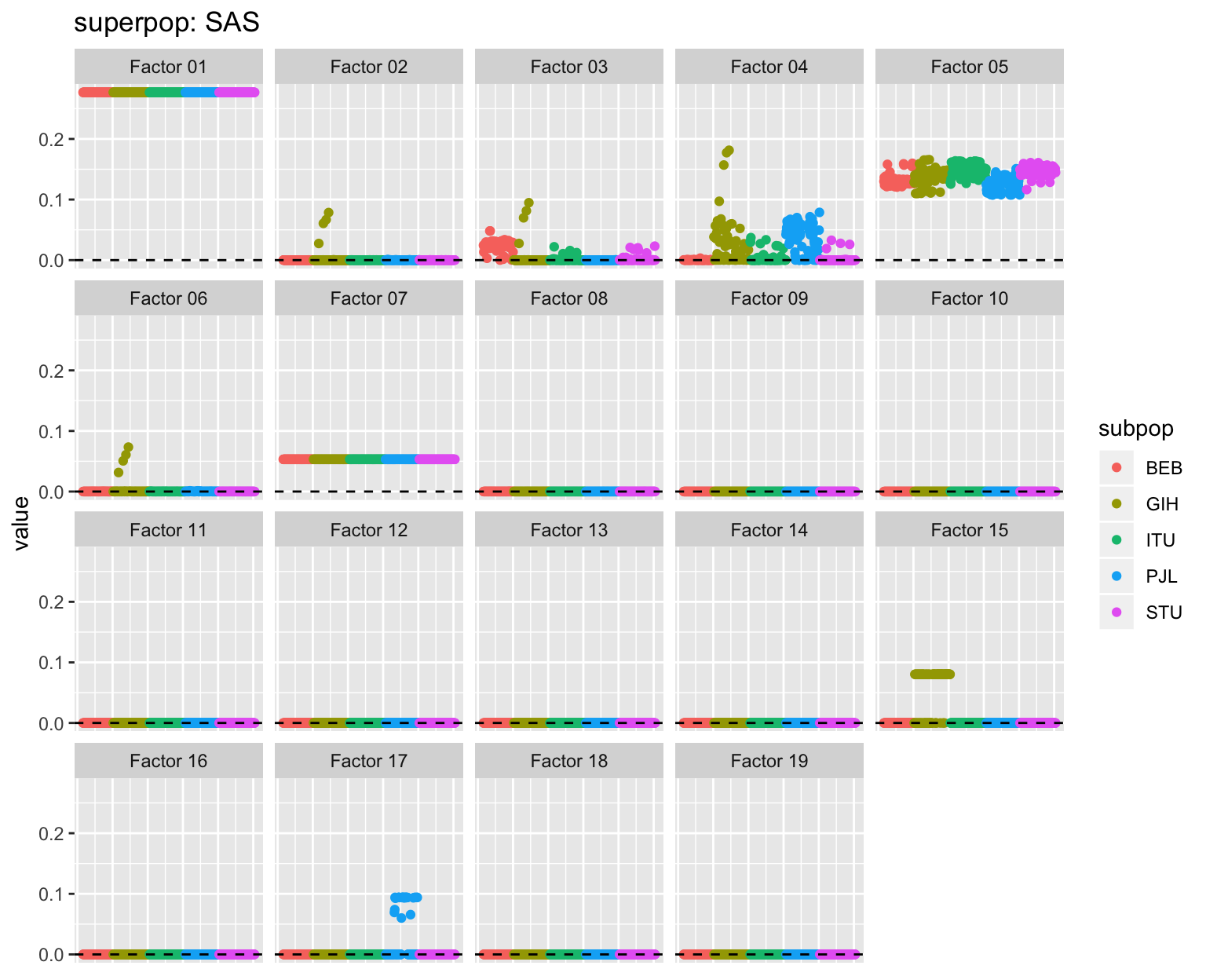
Factor descriptions
Factor 2: The primary African factor, with varying degrees of admixture for African Americans (ASW), African Caribbeans (ACB), and Amerindians. Intriguingly, there is a small degree of admixture for several Iberians (IBR) and Gujarati Indians from Houston (GIH).
Factor 3: The primary East Asian factor. There is a very small amount of admixture for Peruvians (PEL) as well as for a small number of African Americans, Mexicans (MXL), and Finns (FIN). A small degree of admixture is common among Bengali (BEB) and Telugu (ITU) Indians and Sri Lankans (STU), and present but less common among Gujarati Indians.
Factor 4: The primary European factor, with strong contributions from South Asian populations (especially Punjabi and Gujarati) and varying degrees of admixture for African Americans, African Caribbeans, and Amerindians. There is a single Vietnamese (KHV) individual with a nonzero loading: this can probably be ignored.
Factor 5: The primary South Asian factor. There is a single African Caribbean individual with a nonzero loading.
Factor 6: The Amerindian factor, with loadings largest among Peruvians and smallest among Puerto Ricans (PUR). Several African Caribbeans are loaded on this factor, as well as, interestingly, a few Gujarati Indians from Houston.
Factor 7: The out-of-Africa factor. Oddly, it is shared by Mexicans, Peruvians, and most Columbians (CLM), but not by most Puerto Ricans.
Factor 8: Substructure among East Asians: large loadings for the more Southeastern Vietnamese and Chinese Dai populations, modest loadings for some Chinese Han, and mostly zero loadings for Japanese.
Factor 9: A West African factor: large loadings for Gambians (GWD) and modest loadings for the Mende from Sierra Leone (MSL).
Factor 10: Shared by all Amerindian and European populations except for Toscani. I don’t know where this comes from.
Factor 11: A Mediterranean factor, possibly? Shared by Southern European (Iberian and Toscani) and Puerto Rican populations and a large number of Mexicans and Colombians.
Factor 12: A Finnish factor, also present among one or two Utah residents with European ancestry (CEU).
Factor 13: Kenyan.
Factor 14: Japanese.
Factor 15: Common to some, but not all Gujarati Indians from Houston.
Factor 16: Shared among Japanese, most Chinese Han from Beijing, and some Southern Chinese Han.
Factor 17: Common to some, but not all Punjabi.
Factor 18: Shared among the two Nigerian populations (Esan (ESN) and Yoruba (YRI)).
- Factor 19: Shared among most Toscani and Iberians and some British and Utah residents. One Chinese Dai individual has a nonzero loading.
sessionInfo()#> R version 3.5.3 (2019-03-11)
#> Platform: x86_64-apple-darwin15.6.0 (64-bit)
#> Running under: macOS Mojave 10.14.6
#>
#> Matrix products: default
#> BLAS: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRblas.0.dylib
#> LAPACK: /Library/Frameworks/R.framework/Versions/3.5/Resources/lib/libRlapack.dylib
#>
#> locale:
#> [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] forcats_0.4.0 stringr_1.4.0 dplyr_0.8.0.1
#> [4] purrr_0.3.2 readr_1.3.1 tidyr_0.8.3
#> [7] tibble_2.1.1 ggplot2_3.2.0 tidyverse_1.2.1
#> [10] drift.alpha_0.0.11 flashier_0.2.7
#>
#> loaded via a namespace (and not attached):
#> [1] Rcpp_1.0.4.6 lubridate_1.7.4 invgamma_1.1
#> [4] lattice_0.20-38 assertthat_0.2.1 rprojroot_1.3-2
#> [7] digest_0.6.18 truncnorm_1.0-8 R6_2.4.0
#> [10] cellranger_1.1.0 plyr_1.8.4 backports_1.1.3
#> [13] evaluate_0.13 httr_1.4.0 pillar_1.3.1
#> [16] rlang_0.4.2 lazyeval_0.2.2 readxl_1.3.1
#> [19] rstudioapi_0.10 ebnm_0.1-21 irlba_2.3.3
#> [22] whisker_0.3-2 Matrix_1.2-15 rmarkdown_1.12
#> [25] labeling_0.3 munsell_0.5.0 mixsqp_0.3-40
#> [28] broom_0.5.1 compiler_3.5.3 modelr_0.1.5
#> [31] xfun_0.6 pkgconfig_2.0.2 SQUAREM_2017.10-1
#> [34] htmltools_0.3.6 tidyselect_0.2.5 workflowr_1.2.0
#> [37] withr_2.1.2 crayon_1.3.4 grid_3.5.3
#> [40] nlme_3.1-137 jsonlite_1.6 gtable_0.3.0
#> [43] git2r_0.25.2 magrittr_1.5 scales_1.0.0
#> [46] cli_1.1.0 stringi_1.4.3 reshape2_1.4.3
#> [49] fs_1.2.7 xml2_1.2.0 generics_0.0.2
#> [52] tools_3.5.3 glue_1.3.1 hms_0.4.2
#> [55] parallel_3.5.3 yaml_2.2.0 colorspace_1.4-1
#> [58] ashr_2.2-51 rvest_0.3.4 knitr_1.22
#> [61] haven_2.1.1